CrusTF FAQ
Searching Transcription Factors
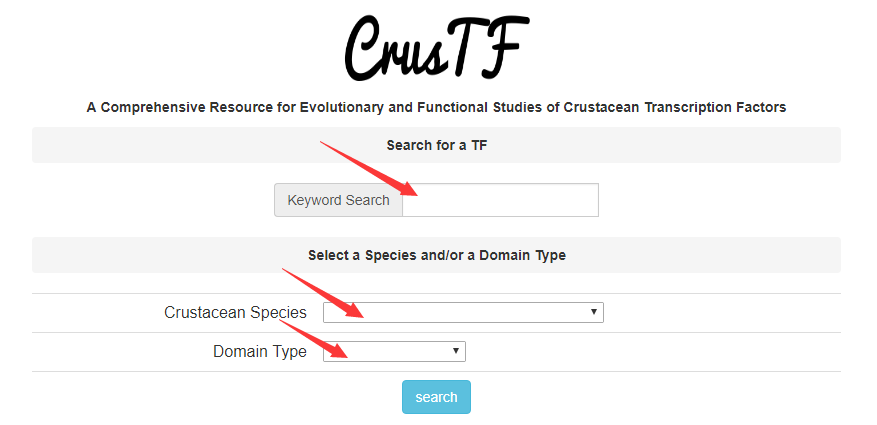
In home page, there are two ways to search a TF. Enter the name of TF in “Keyword Search”. You can enter the FULL name or parts of it, but two or more keywords are illegal. Use combo boxes below to select a species and/or a DNA binding domain type to narrow down the search. You can also leave the “Keyword Search” blank to list all TFs with a certain domain type in a certain species. And matching list will be given in result page. Each TF name is linked to a page of TF information.
Example:
Input "AgTHAP4" or a substring of it "THAP4"
Press "search", we will see
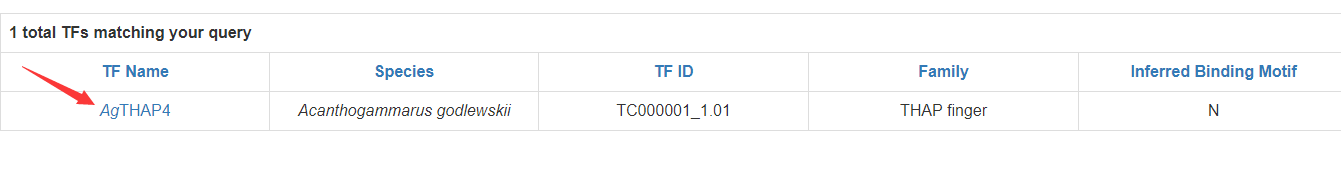
In the table, each column means:
TF Name: name of the TF.
Species: scientific name of the species.
TF ID: TF ID in CrusTF.
Family: TF family the TF belongs to.
Inferred Binding Motif: whether the TF has inferred binding motif. “Y”: inferred binding motif is available; “N”: inferred binding motif is not available.
Each row of ‘TF Name’ is linked to the page of TF information: Sequences of CDS, protein and binding domain of the TF could be found in this page.
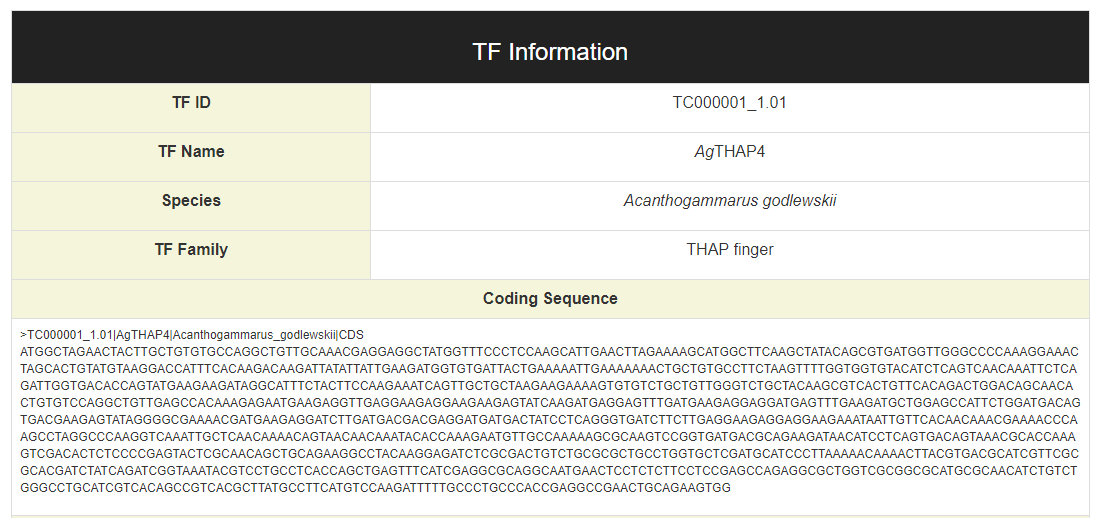
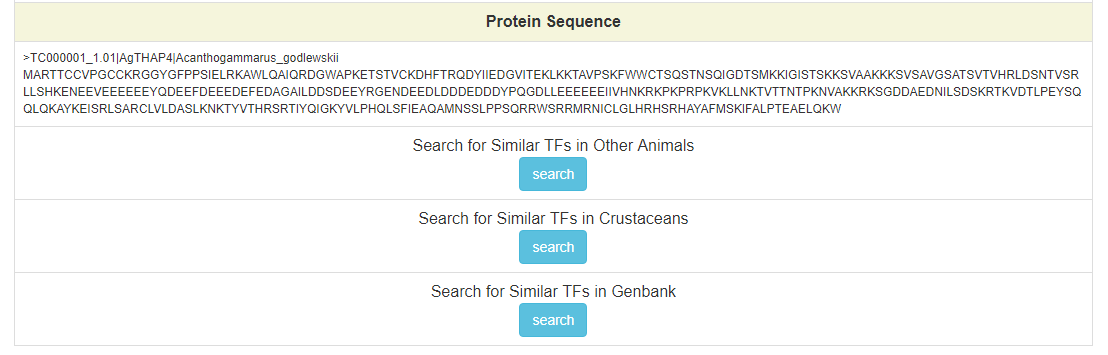
Click on the “search” buttons in the middle of this page, CrusTF will search similar TF in other animals (upper button), crustaceans (middle button) or (lower button), and get a list of similar TFs:
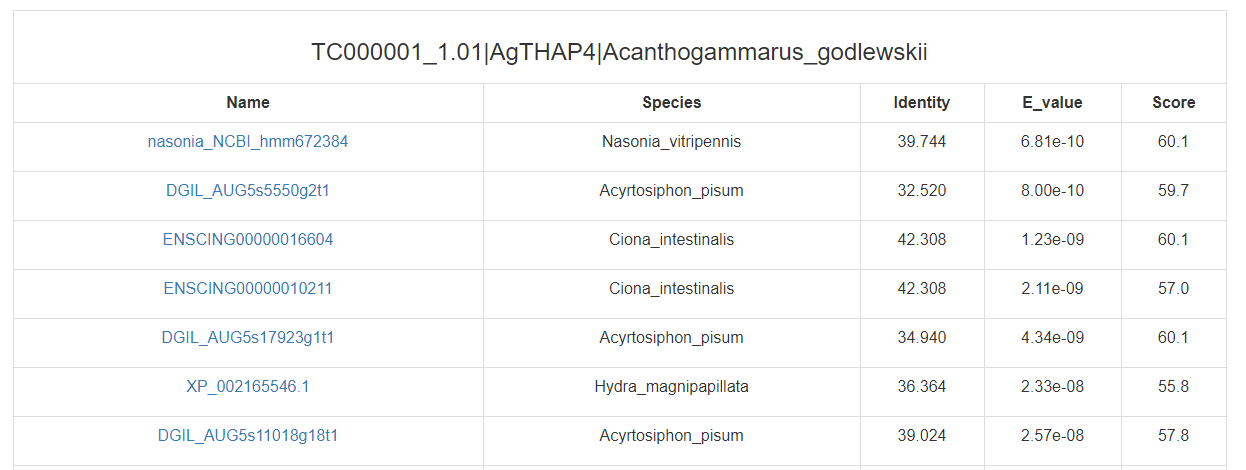
(in other animals)
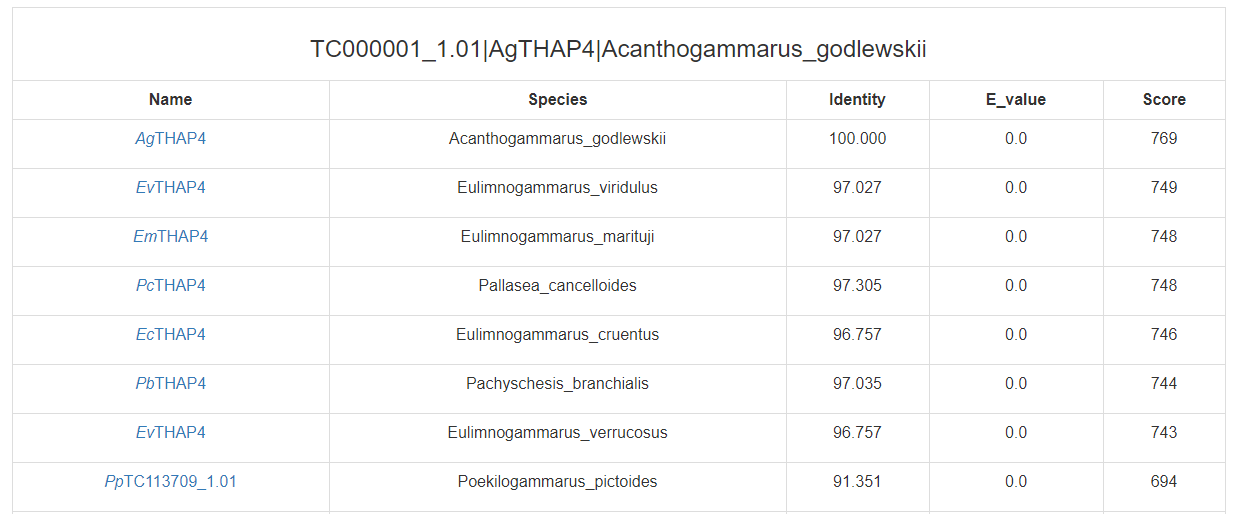
(in crustaceans)
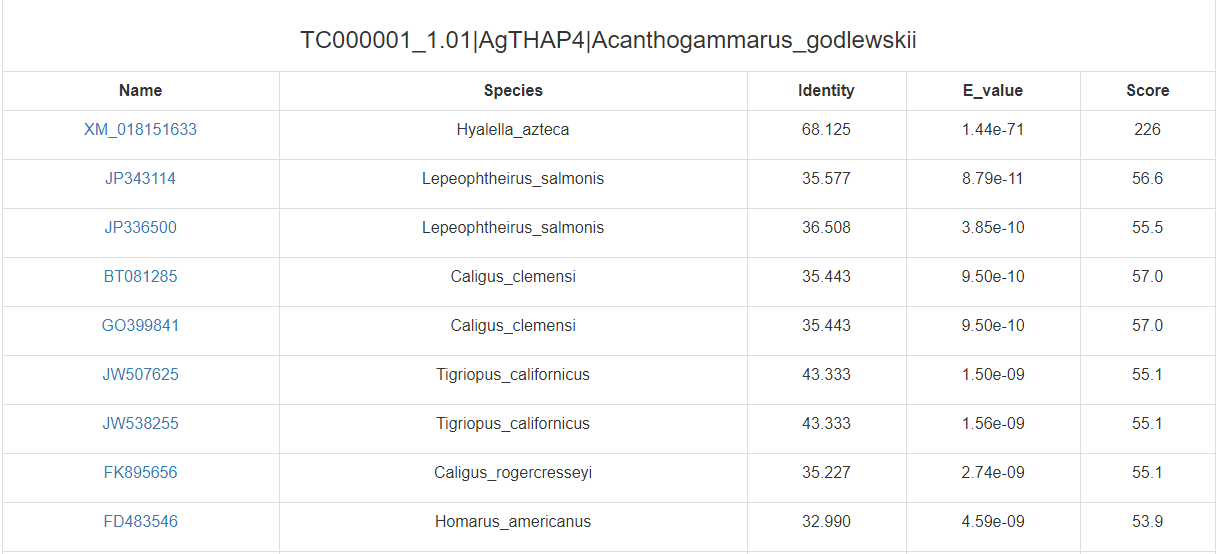
(in ncbi GeneBank)
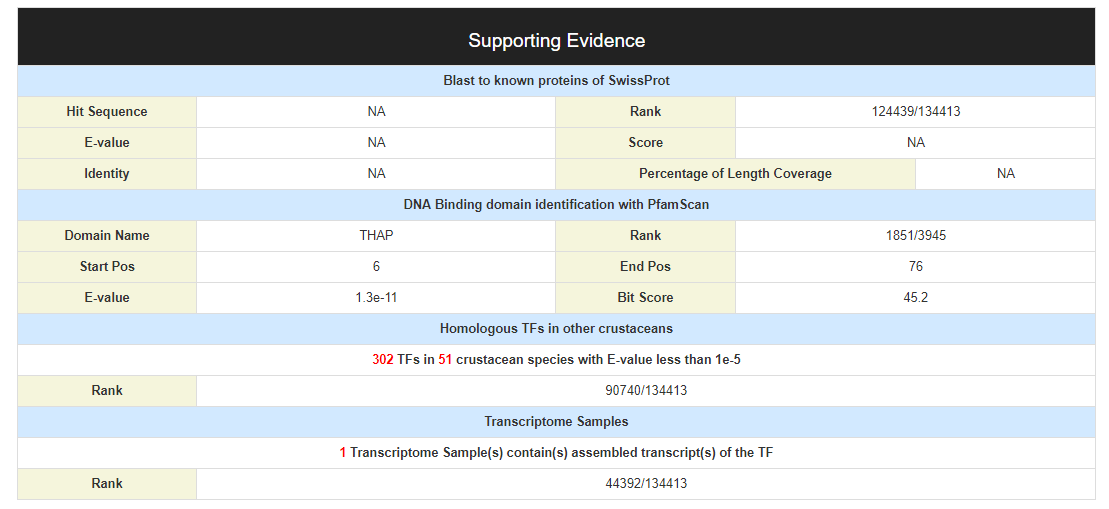
The confidence of predicted TF was estimated with several evidences: 1) the percentage of the predicted TF that matches the top hit when Blast to the protein database SwissProt, the E-value, Blast score and identity 2) the bit-score and E-value of PfamScan 3) the number of homologs found in other crustacean species 4) the number of transcriptome samples in which the TF was detected For example :(NA means no evidence to support)
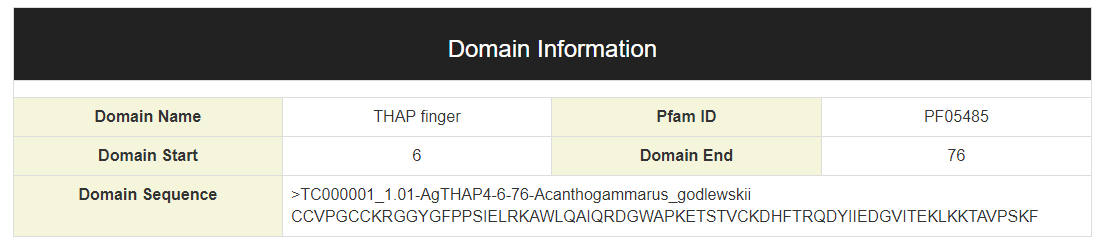
The domain information is shown below
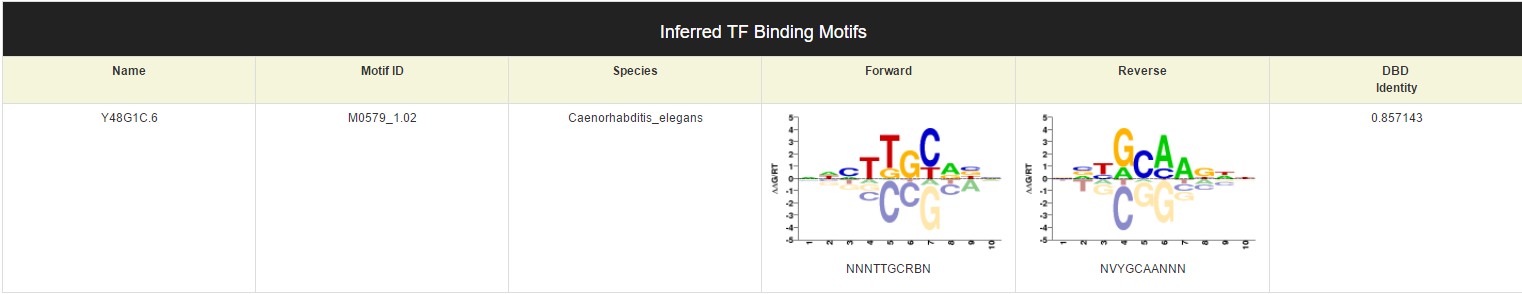
If this TF have inferred binding motif, the last part of this page will shows the motif information.
Trees of DNA Binding Domains
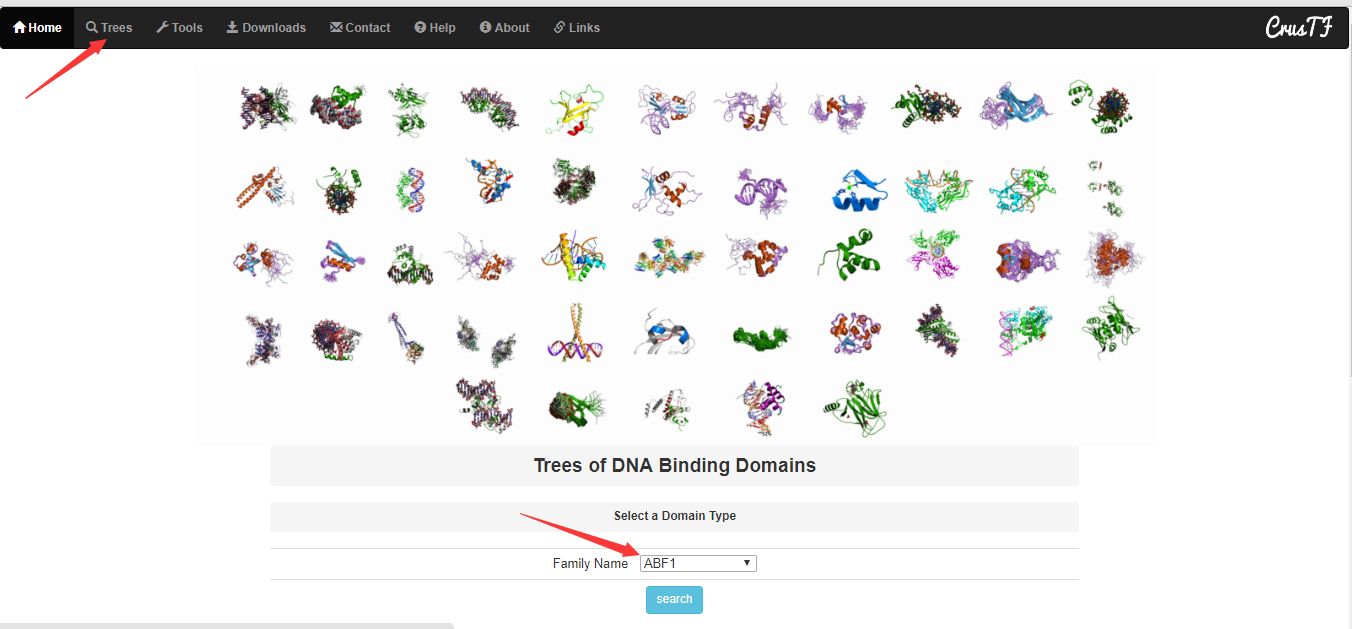
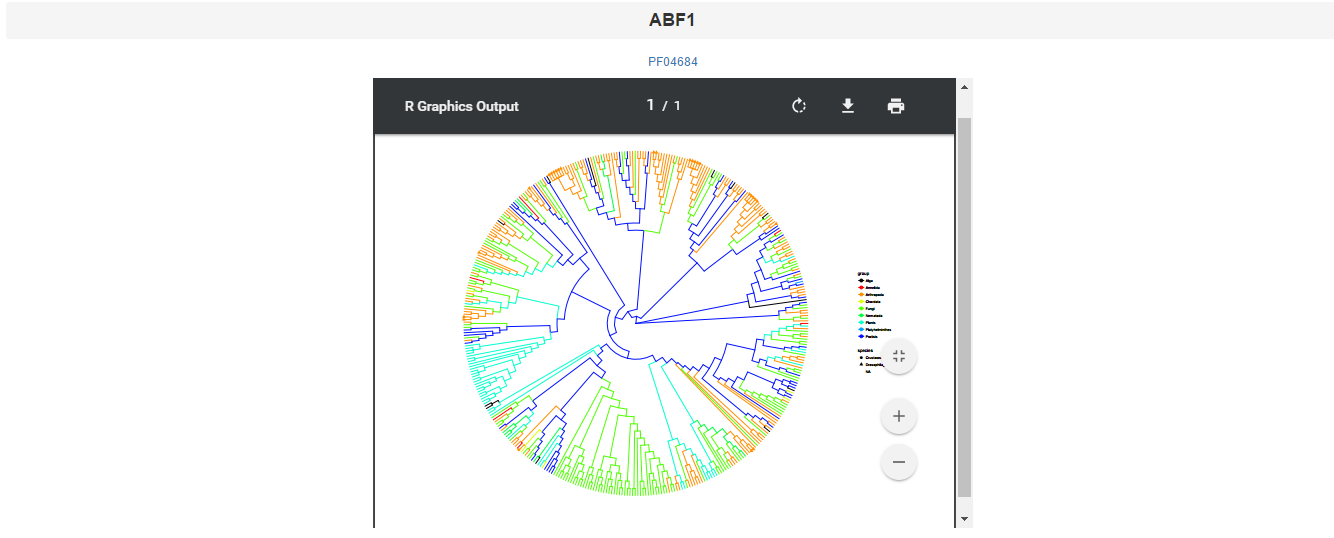
This page offers a way to browse the trees of DNA binding domain. Select a domain type the tree figure will show as:
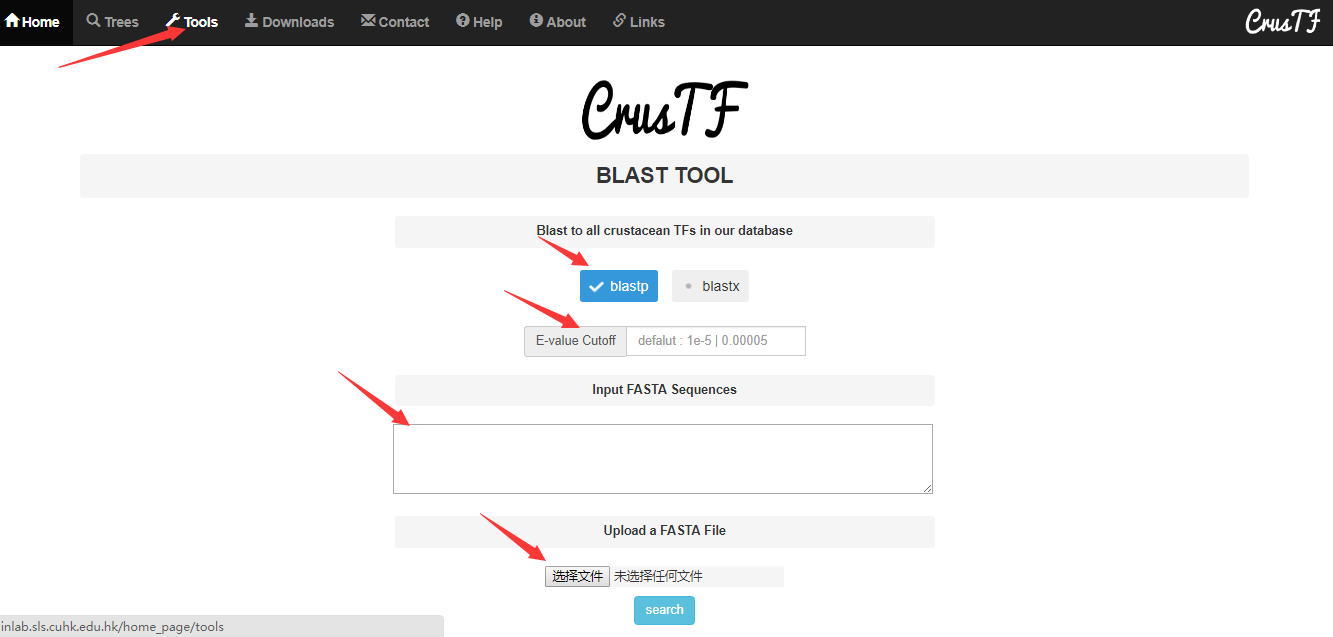
Blast tools
CrusTF provides a portal to blast our database to compare a TF given by user to TFs in our database. Users can paste fasta sequences or upload a fasta file. Users can use blastp to search our TF protein sequences if their inputted sequences are proteins, or use blastx if their inputted sequences are DNAs. E-value cutoff could be specified by the users (default 1e-5).
Example:
Use AgTHAP4 as an example.
This is the protein sequence of AgTHAP4.
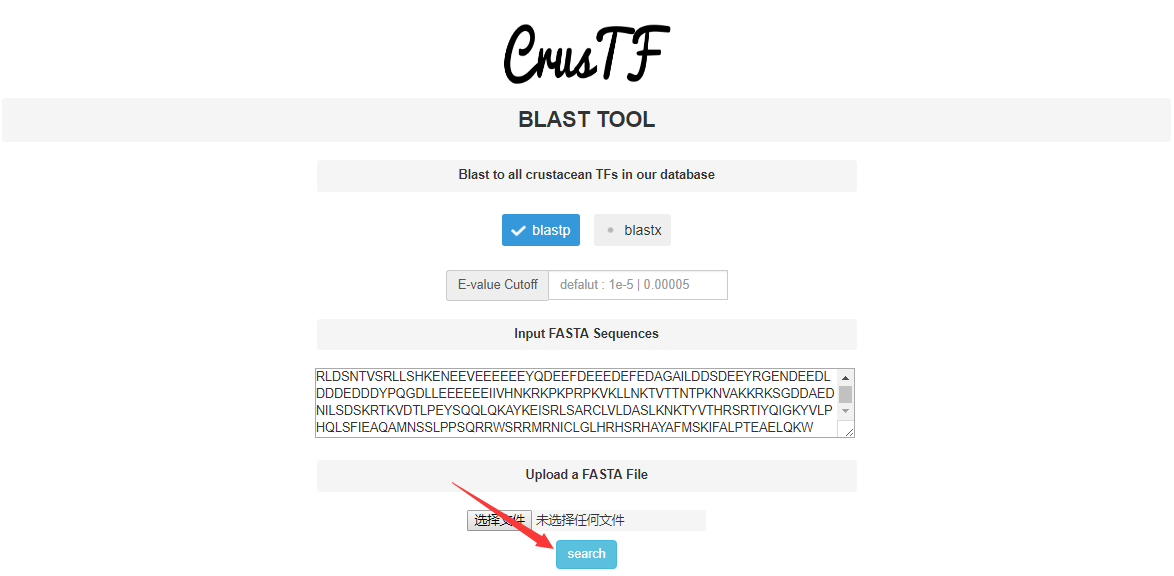
Input in textbox and click on "search"
Blastp results will be listed as:
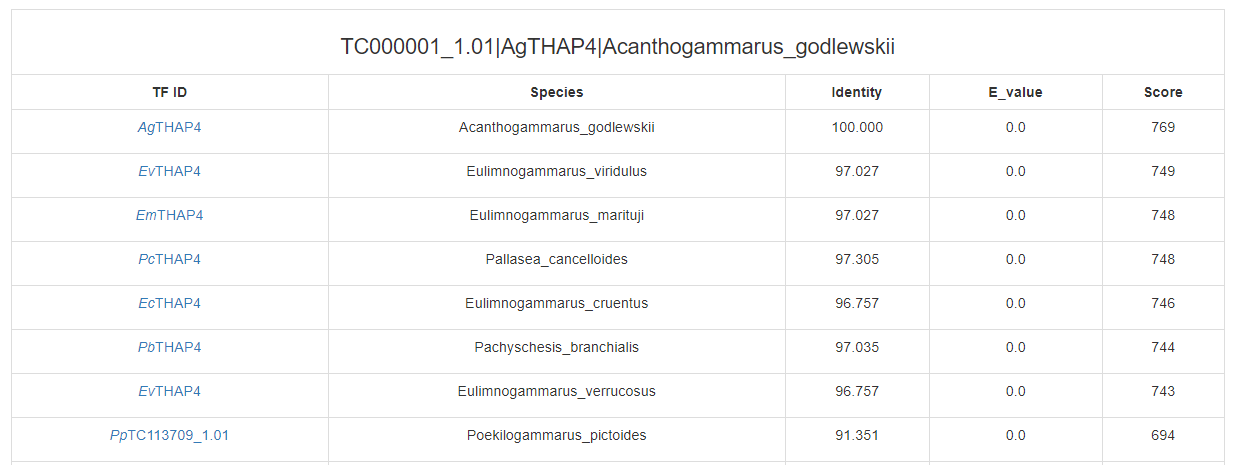
Each matched TF will link to its TF information page.
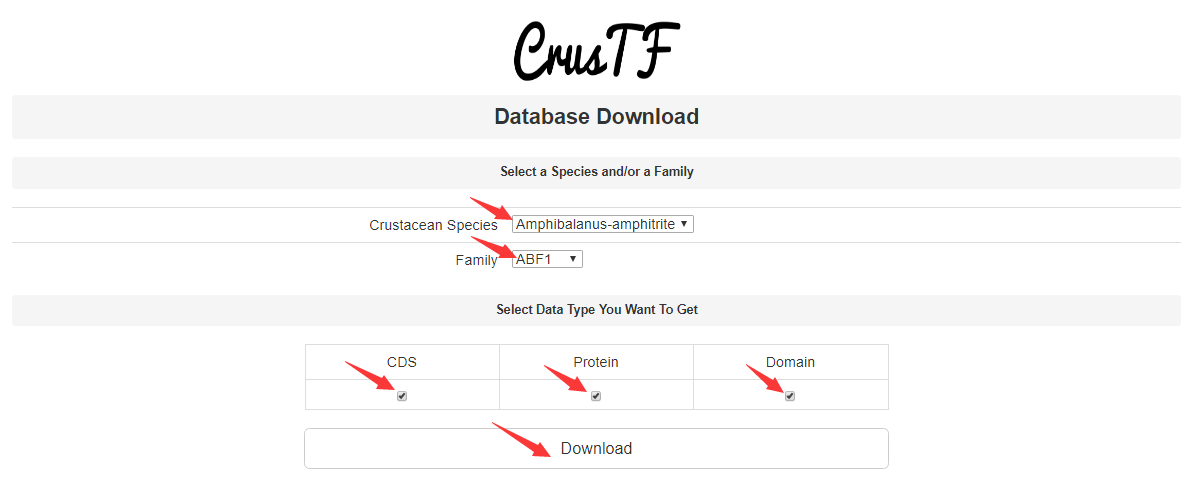
Download Database
CrusTF allows users to download the sequences of crustacean TFs. Users can select the species and TF family they want and download CDS, protein, or/and domain sequences. Users can also download sequences of a certain TF family in all species by leaving the “Crustacean Species” blank and selecting a family, download sequences of all TF families in a certain species by leaving the “Family” blank and selecting a species, or download all sequences by leaving both blank.
Example: