CrusTF: A Comprehensive Resource for Evolutionary and Functional Studies of Crustacean Transcription Factors
Crustacea, the second largest subphylum of Arthropoda, includes species of major ecological and economic importance, such as crabs, lobsters, crayfishes, shrimps and barnacles. With the rapid development of crustacean aquaculture and biodiversity loss, understanding the gene regulatory mechanisms of growth, reproduction and development of crustaceans is crucial to both aquaculture development and biodiversity conservation of this group of organisms. In these biological processes, transcription factors (TFs) play a vital role in regulating gene expression. However, crustacean TFs are still largely unknown. Because the lack of complete genome sequences of most crustacean species, the current TF databases derived from genome sequences contain TF information for only a few crustacean species, and are insufficient to elucidate the transcriptional diversity of such a large animal
CrusTF fills the knowledge gap of transcriptional regulatory systems in crustaceans by exploring publicly available and newly sequenced transcriptomes more than 200 crustacean species and identifying 131,941 and 8,502 TFs of crustacean species from transcriptomes and Genbank from 63 TF familes respectively. CrusTF features three categories of information: sequence, function and evolution of crustacean TFs. Given the importance of TF information in evolutionary and functional studies on transcriptional regulatory systems of crustaceans, this database will constitute a key resource for the research community of crustacean biology and evolutionary biology. Moreover, CrusTF serves as a model of TF database derived from transcriptome data. Similar approach could be applied to other groups of organisms, for which transcriptomes are more readily available than genomes.
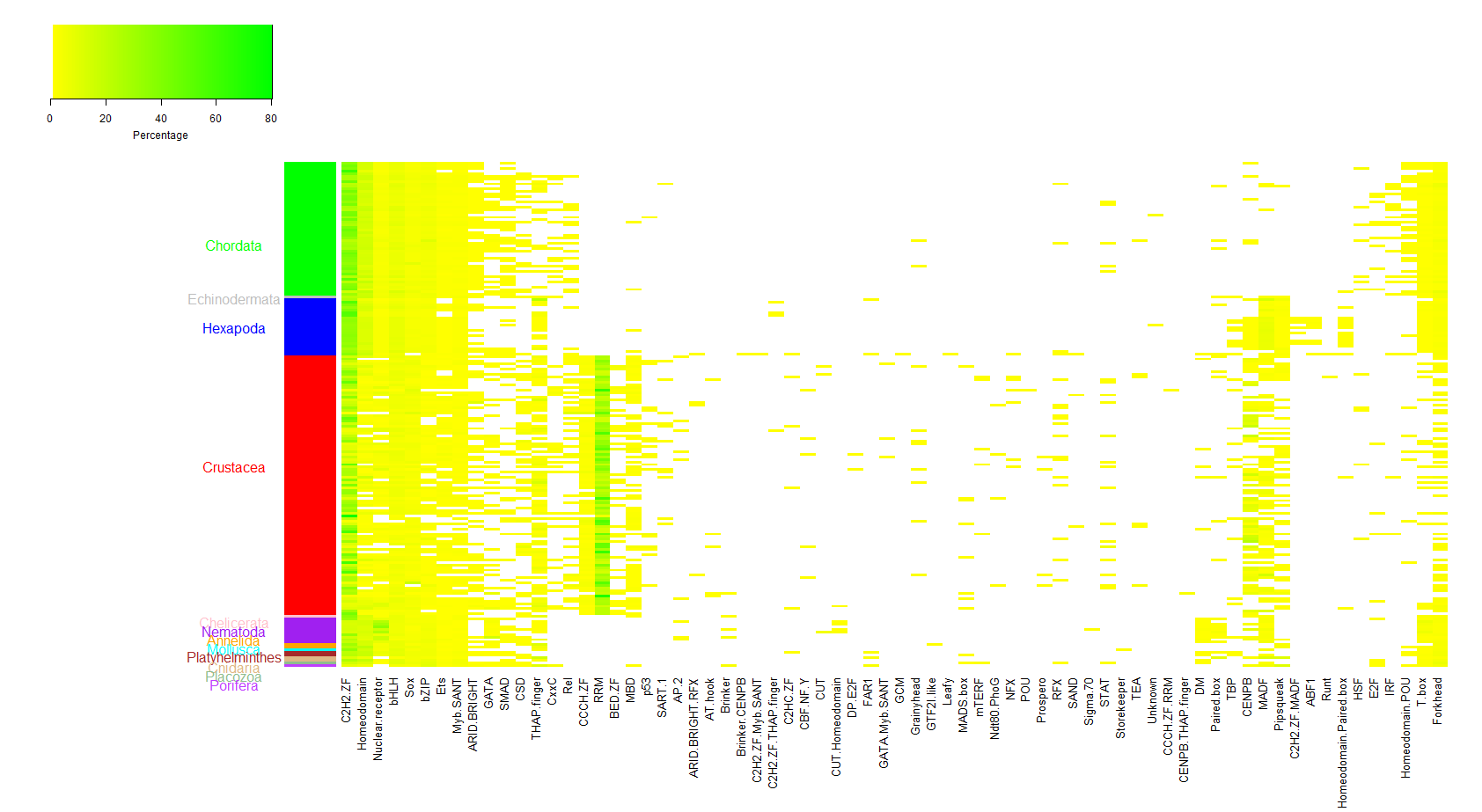
Figure 1. TF families in crustaceans compared to those in other animals. Colors in the figure show the percentage of TFs in each TF family over all predicted TFs in a species (white, <1%; yellow to green, 1-100%). Each row is a species and each column is a TF family. Side bar highlights the taxa. Many TF families on the left that are prevalently detected in metazoans were detected in most crustaceans. Several TFs families, such as families with Zinc finger CCCH domain (CCCH ZF) and BED zinc finger (BED ZF), show extensive expansion in crustaceans. Some TF families with distinct DBD combinations may represent putative TFs unique to this animal group and have not been characterized in other TF databases.
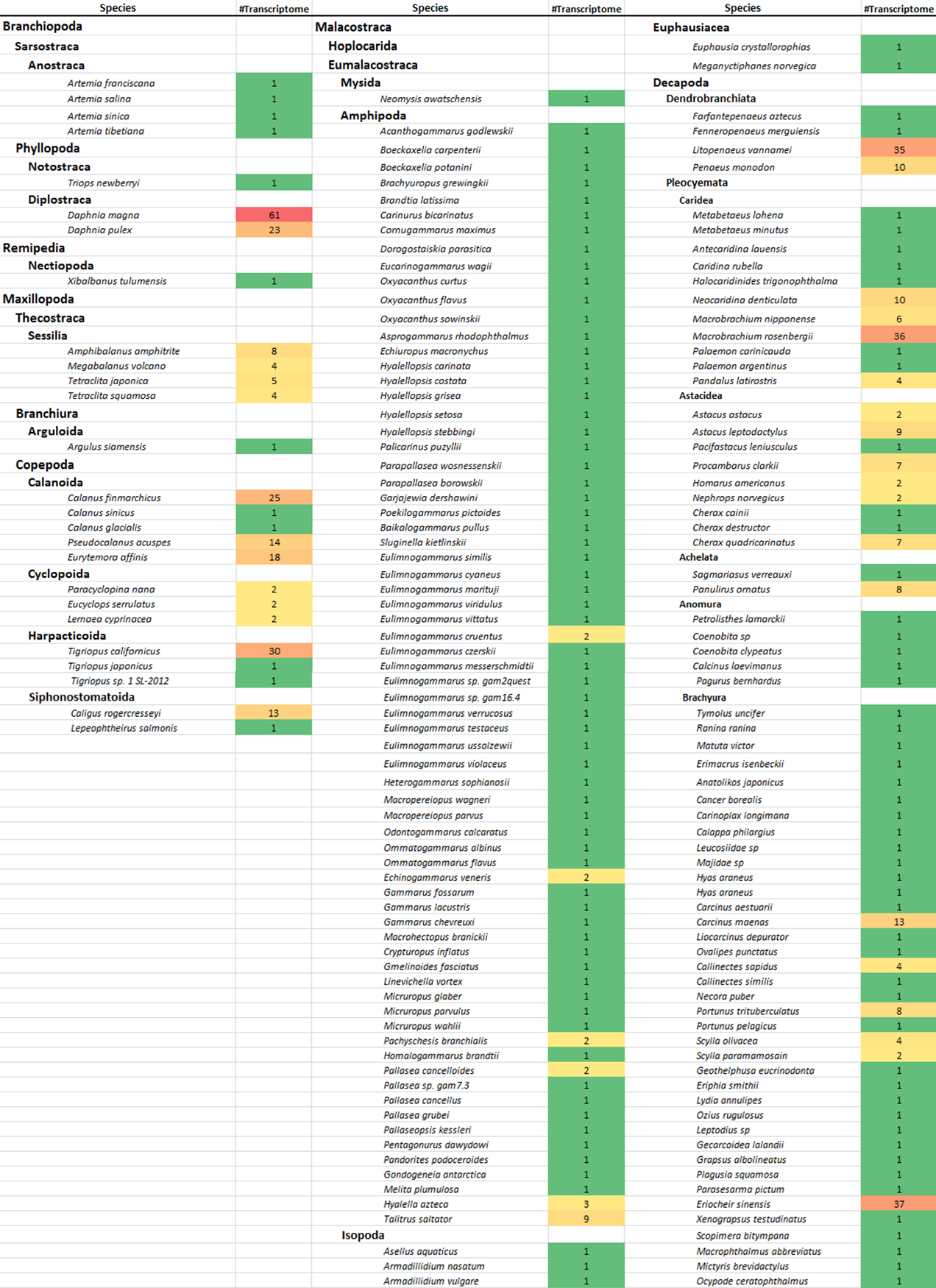
Figure 2. Collection of crustacean transcriptomes in CrusTF.Transcriptomes were collected from public databases, short read archive (SRA) and transcriptome shotgun assembly (TSA). Crustacean genes, including those derived from low-throughput experiments, were also downloaded from Genbank and included in this database. CrusTF covers over 200 crustacean species belonging to 15 orders.
This work was supported by a grant from the National Natural Science Foundation of China (NSFC) (project no. 41606143), a Direct Grant for Research from The Chinese University of Hong Kong (project no. 4053187), a grant from Collaborative Research Fund of the Research Grants Council, Hong Kong SAR Government (project no. C4042-14G), and grants from NSFC (project no. 11601343) and Natural Science Foundation of Guangdong (project no. 2016A030310038)