Searching for Nucleic acid Binding Proteins (NBPs)
Click the ‘Tools’ button on the top bar. Select "Search"
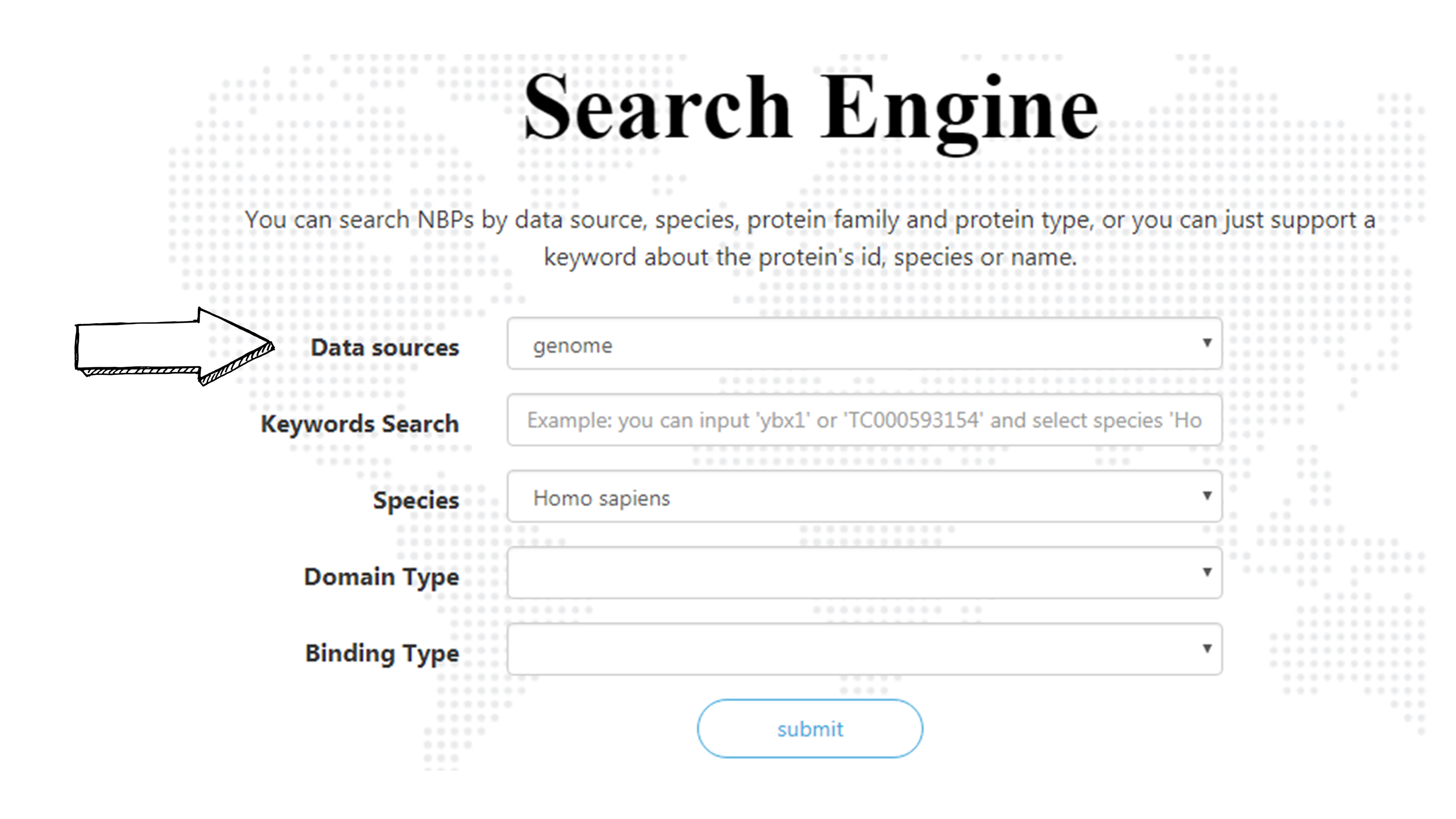
Select the source of your interested NBP originated from: Genomes or Transcriptomes
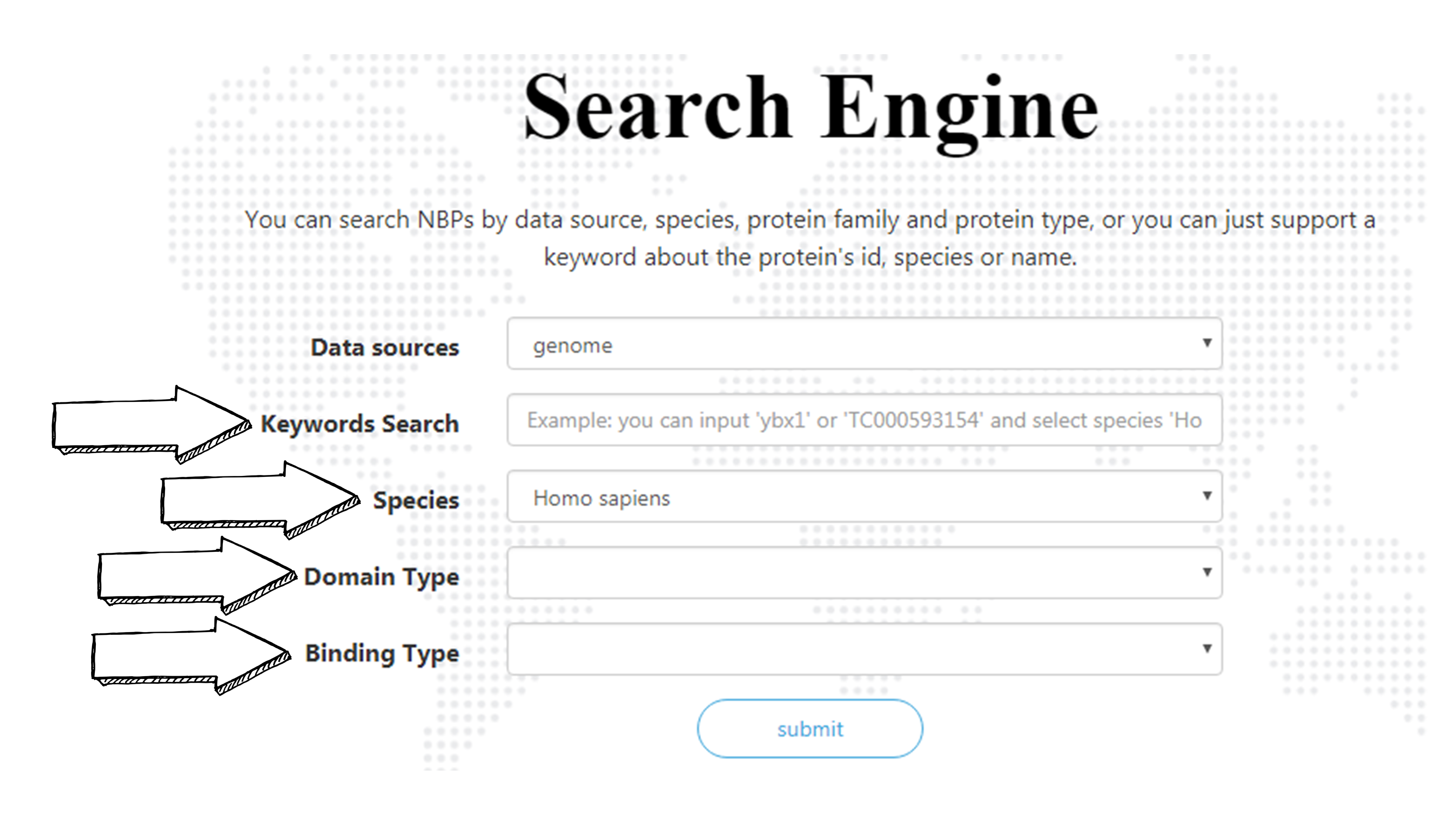
There are two ways to search a NBP. Enter the name or id of NBP in “Keyword Search”. You can enter the FULL name or parts of it, but two or more keywords are illegal. Use combo boxes below to select a species, nucleic acid binding type and/or a nucleic acid binding domain type to narrow down the search. You can also leave the “Keyword Search” blank to list all NBPs with a certain domain type in a certain species. And matching list will be given in result page. Each NBP name is linked to a page of NBP information.
Example:
Input "ybx1" or a substring of it "ybx", selecting Homo_sapiens in the species selection box and then press submit, it shall return with a search result list.
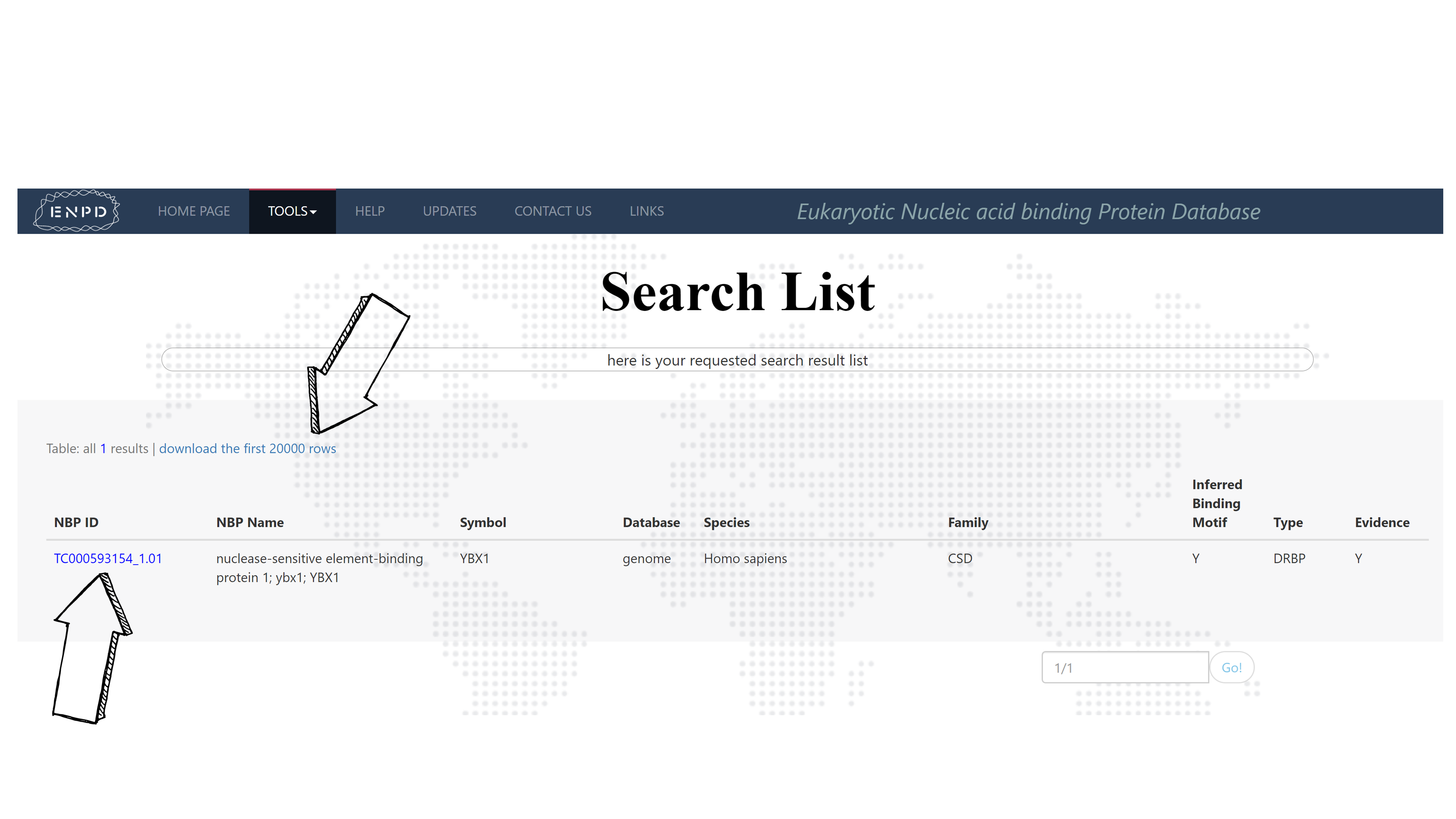
Press the button on top of the result list to download the first 20000 of the list. While pressing the TF ID of the NBP you are interested would link you to an NBP information page as follow.
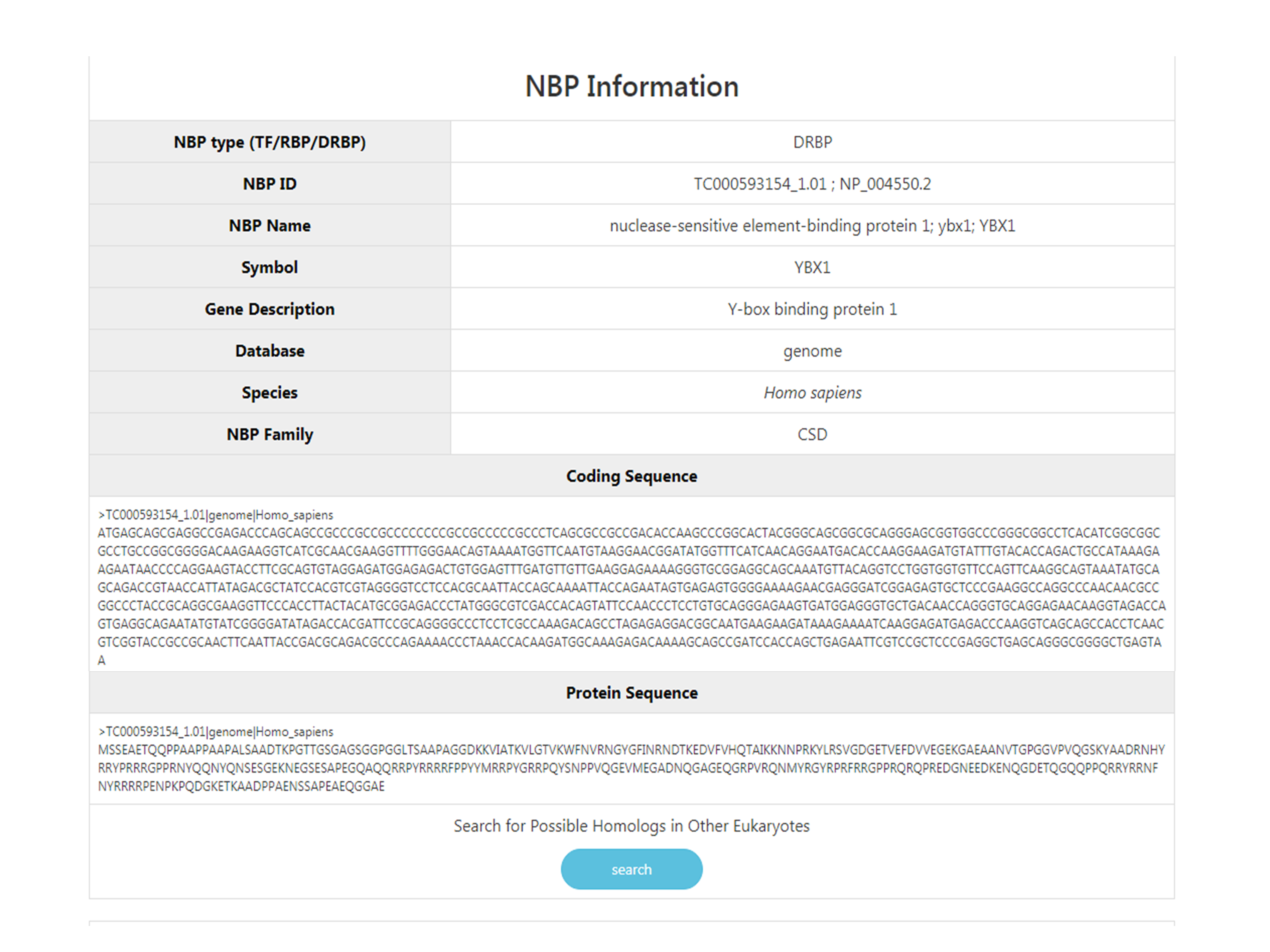
This section provides basic NBP information such as NBP name, gene symbol, NBP ID, data origin (whether it is from genome or transcriptome), the specie it belongs to and its coding and protein sequences. There is also a button where you can try to blast this protein with all other NBPs in this database real time in order to search for possible homologs in other eukaryotes.
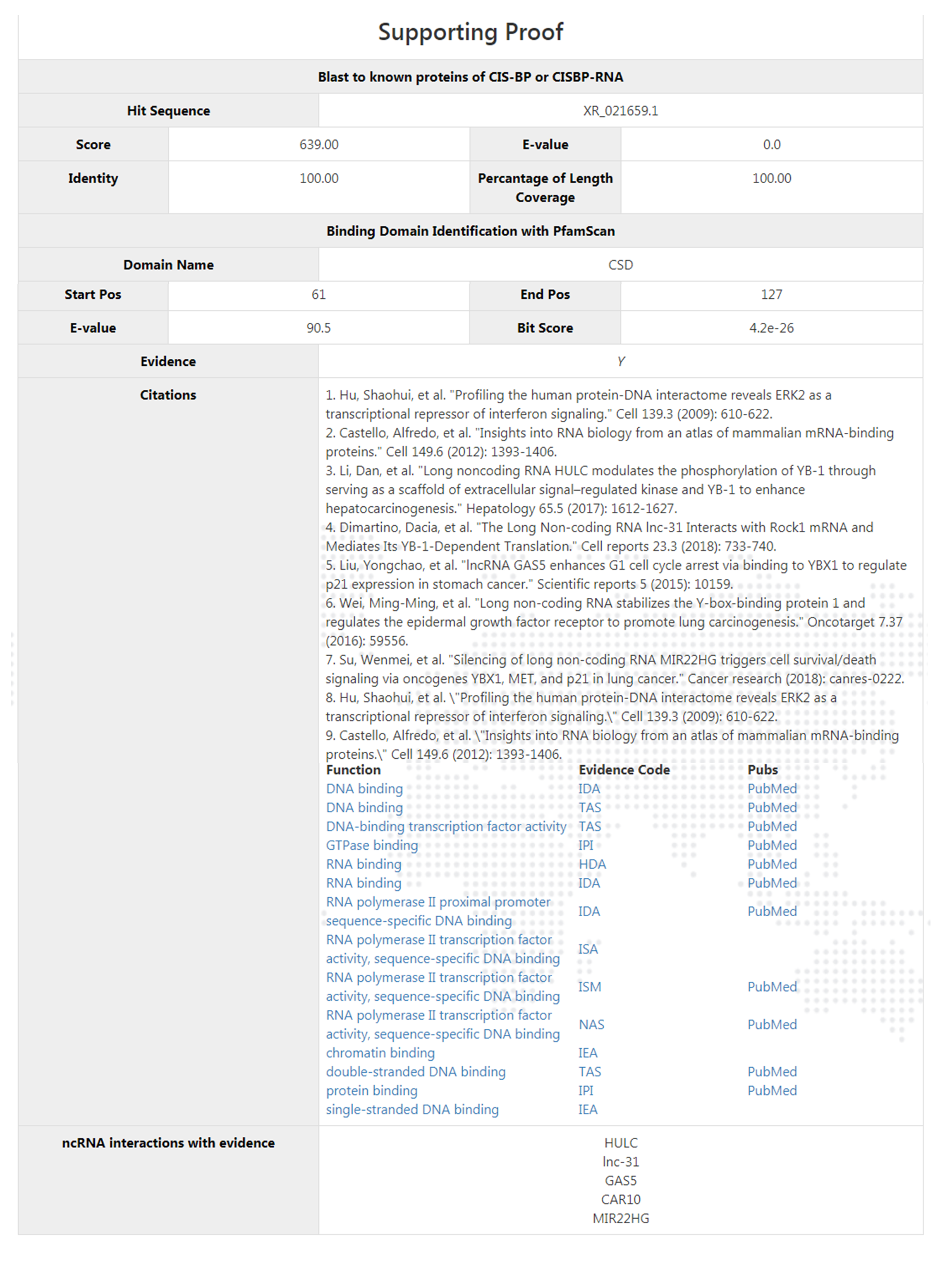
Right down below comes with the supporting proof section. Here it gives the best hit blast result scores of this protein blasted towards proteins in CIS-BP and CISBP-RNA as well as the PfamScan identification scores. Functionality citations and annotations are also provided. The users would be sufficient to make a judgment of the confident level of the query based on such information.
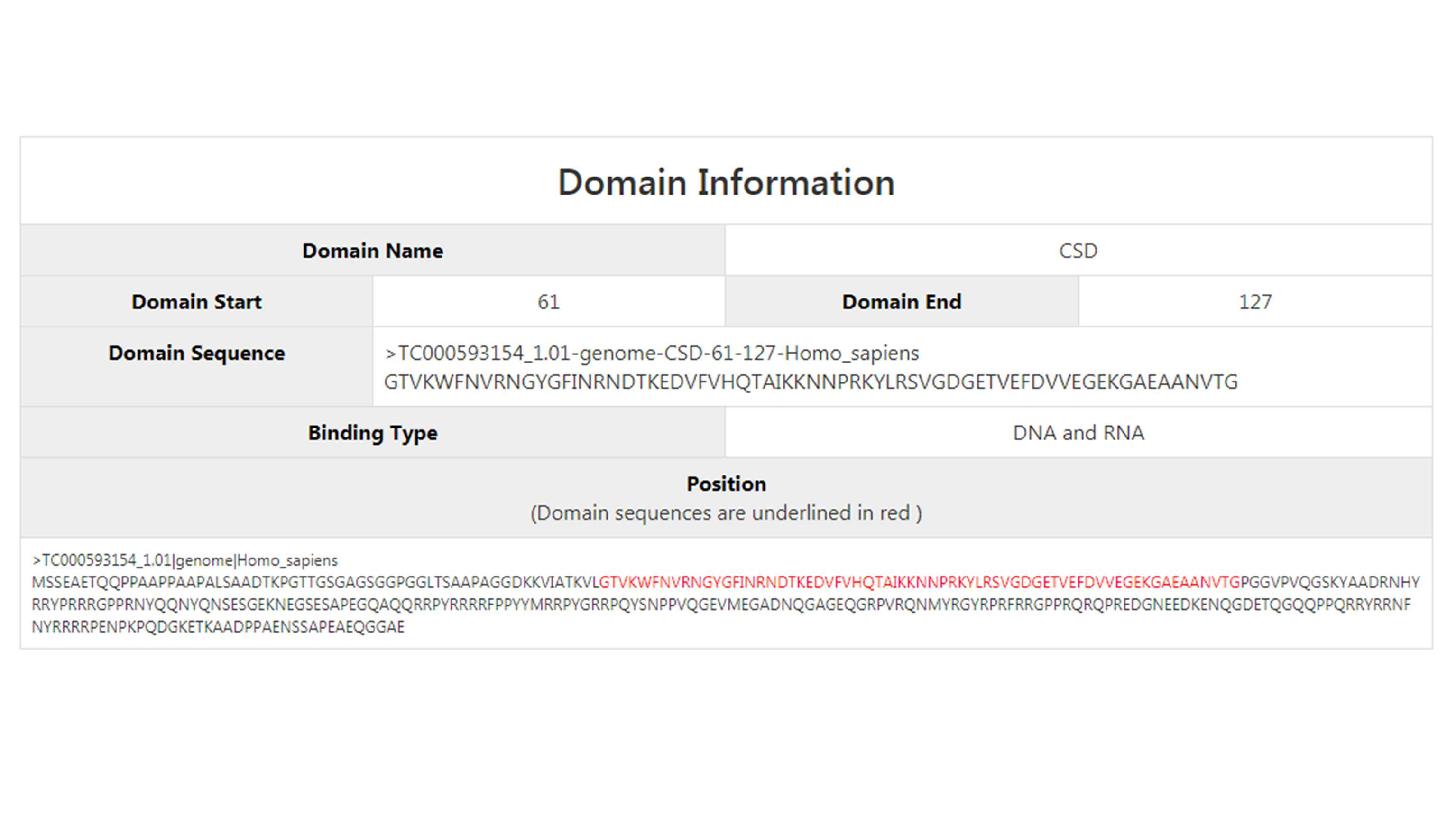
While the domain information is shown further down here. Here it shows the name of the domain, the domain start and end sites in the protein, the domain sequence, the nucleic acid that it can possibly bind to and its position in the protein is shown in red.
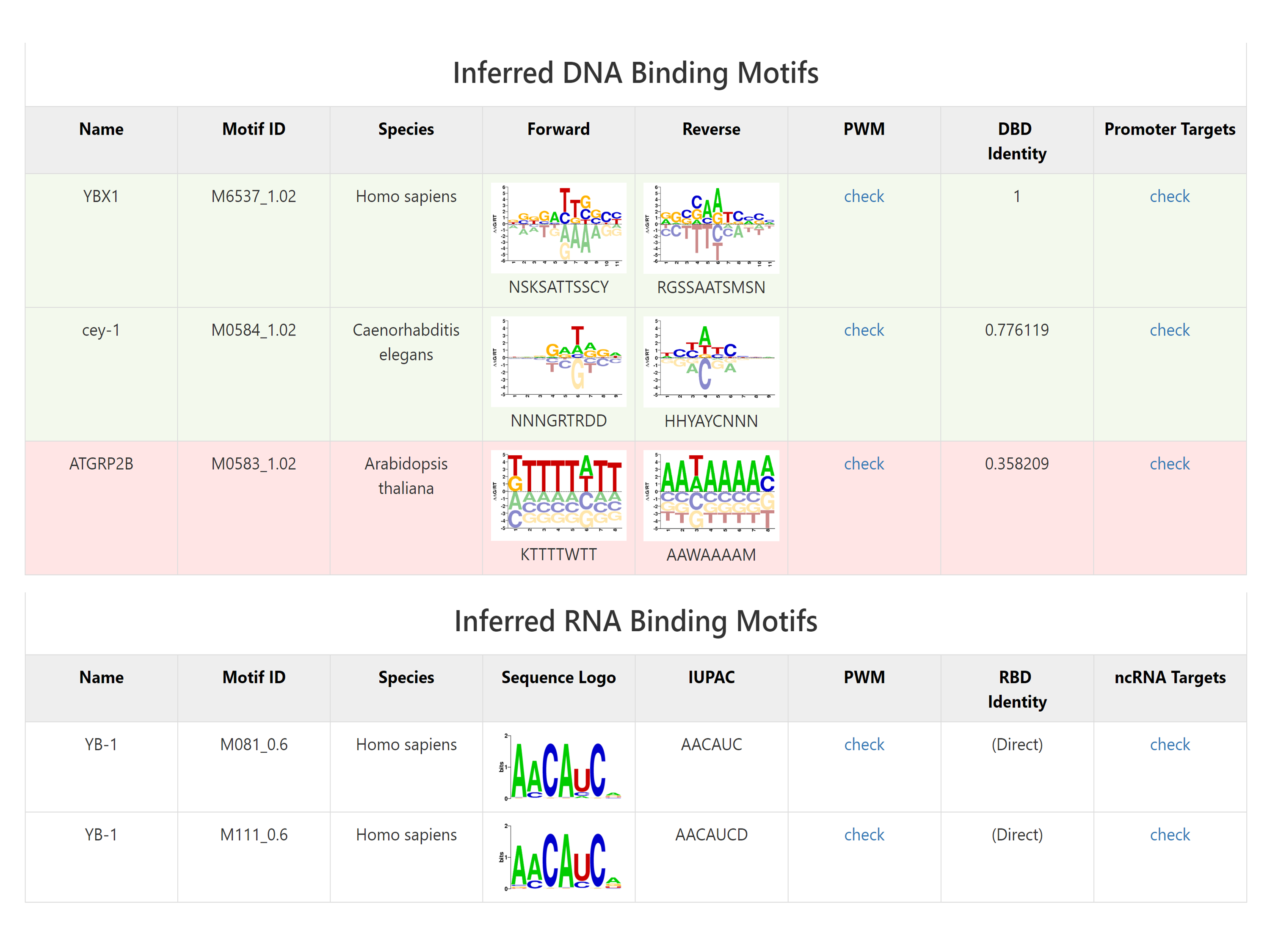
If this NBP have inferred binding motif, the following part will show the motif information. The domain identity cutoff scores for motif inferring is listed as follow.
| Pfam ID | Identity cutoff |
|---|---|
| PF00010 | 0.6 |
| PF00013 | 0.7 |
| PF00035 | 0.7 |
| PF00046 | 0.7 |
| PF00076 | 0.7 |
| PF00096 | 0.7 |
| PF00098 | 0.7 |
| PF00105 | 0.7 |
| PF00157 | 0.7 |
| PF00170 | 0.55 |
| PF00172 | 0.75 |
| PF00178 | 0.5 |
| PF00249 | 0.875 |
| PF00250 | 0.4 |
| PF00292 | 0.7 |
| PF00313 | 0.7 |
| PF00319 | 0.7 |
| PF00320 | 0.7 |
| PF00352 | 0.7 |
| PF00447 | 0.7 |
| PF00505 | 0.925 |
| PF00536 | 0.7 |
| PF00554 | 0.525 |
| PF00575 | 0.7 |
| PF00605 | 1 |
| PF00641 | 0.7 |
| PF00642 | 0.7 |
| PF00649 | 0.7 |
| PF00751 | 0.85 |
| PF00806 | 0.7 |
| PF00847 | 0.7 |
| PF00853 | 0.7 |
| PF00870 | 0.7 |
| PF00907 | 0.6 |
| PF01285 | 0.7 |
| PF01342 | 0.775 |
| PF01388 | 0.65 |
| PF01422 | 0.7 |
| PF01429 | 0.7 |
| PF01530 | 0.7 |
| PF01698 | 0.7 |
| PF02008 | 1 |
| PF02037 | 0.7 |
| PF02042 | 0.7 |
| PF02045 | 0.7 |
| PF02178 | 0.4 |
| PF02200 | 0.7 |
| PF02257 | 0.25 |
| PF02319 | 0.85 |
| PF02362 | 0.7 |
| PF02365 | 0.65 |
| PF02376 | 0.7 |
| PF02536 | 0.7 |
| PF02701 | 1 |
| PF02864 | 0.7 |
| PF02892 | 0.7 |
| PF02946 | 0.7 |
| PF03101 | 0.7 |
| PF03106 | 0.35 |
| PF03110 | 0.625 |
| PF03165 | 0.7 |
| PF03195 | 0.7 |
| PF03299 | 0.7 |
| PF03343 | 0.7 |
| PF03514 | 0.7 |
| PF03615 | 0.5 |
| PF03634 | 0.225 |
| PF03638 | 0.7 |
| PF03859 | 0.7 |
| PF04001 | 0.7 |
| PF04146 | 0.7 |
| PF04218 | 0.7 |
| PF04383 | 0.8 |
| PF04504 | 0.7 |
| PF04516 | 0.7 |
| PF04542 | 0.7 |
| PF04640 | 0.7 |
| PF04684 | 0.7 |
| PF04689 | 0.7 |
| PF04769 | 0.7 |
| PF04873 | 0.35 |
| PF05044 | 0.7 |
| PF05224 | 0.7 |
| PF05225 | 0.85 |
| PF05383 | 0.7 |
| PF05485 | 0.975 |
| PF06217 | 0.7 |
| PF07528 | 0.7 |
| PF08536 | 0.7 |
| PF08731 | 0.7 |
| PF08781 | 0.7 |
| PF08879 | 0.7 |
| PF09197 | 0.7 |
| PF09271 | 0.7 |
| PF09607 | 0.7 |
| PF10283 | 0.7 |
| PF10416 | 0.7 |
| PF10545 | 0.65 |
| PF12550 | 0.7 |
| Unknown | 0.7 |
And there are links linking to the DNA or ncRNA target list of the motifs.
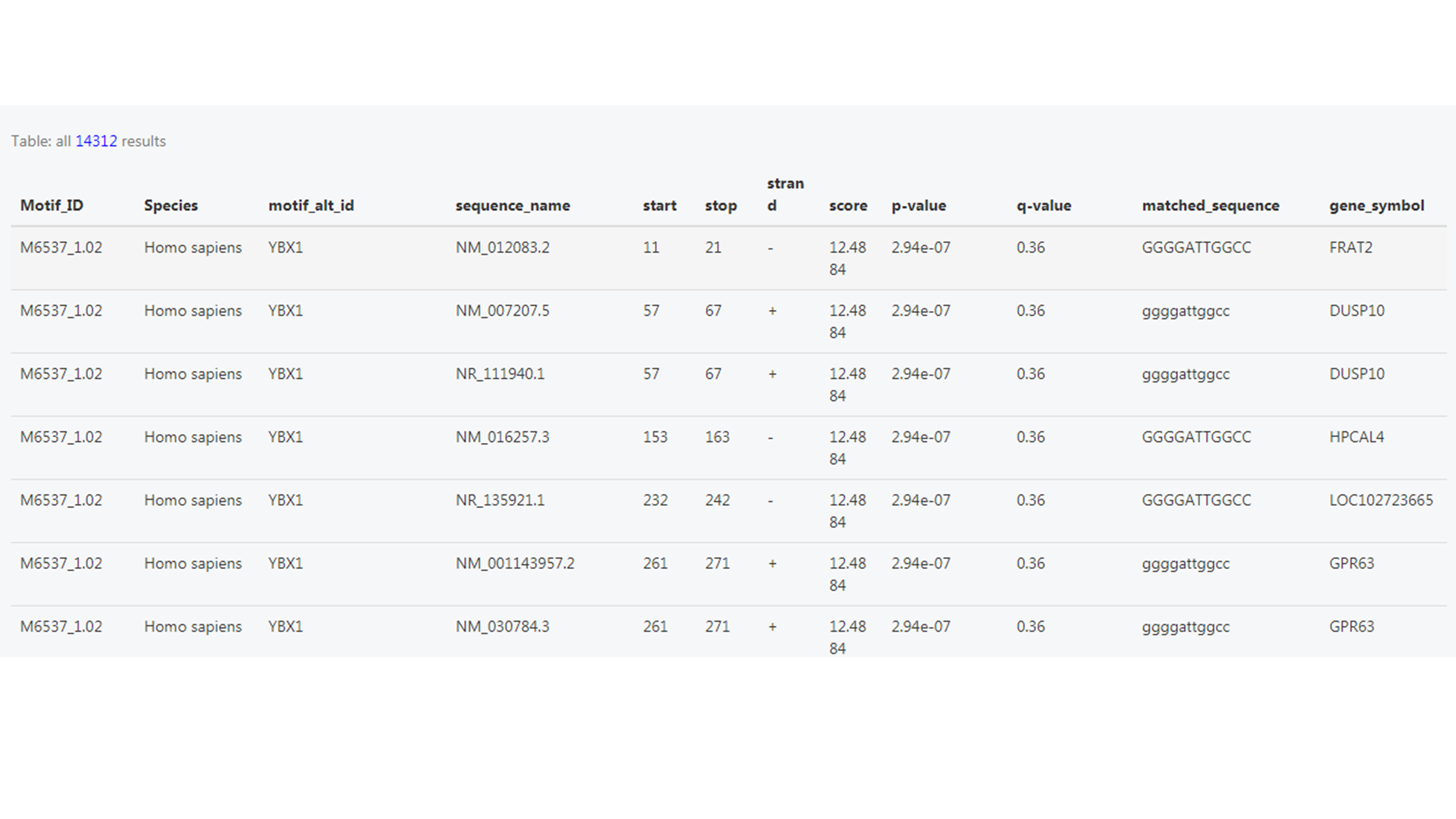
Where it shows the sequence name (ID of the target sequence), predicted scores, matched sequence and the gene symbol of the targets.
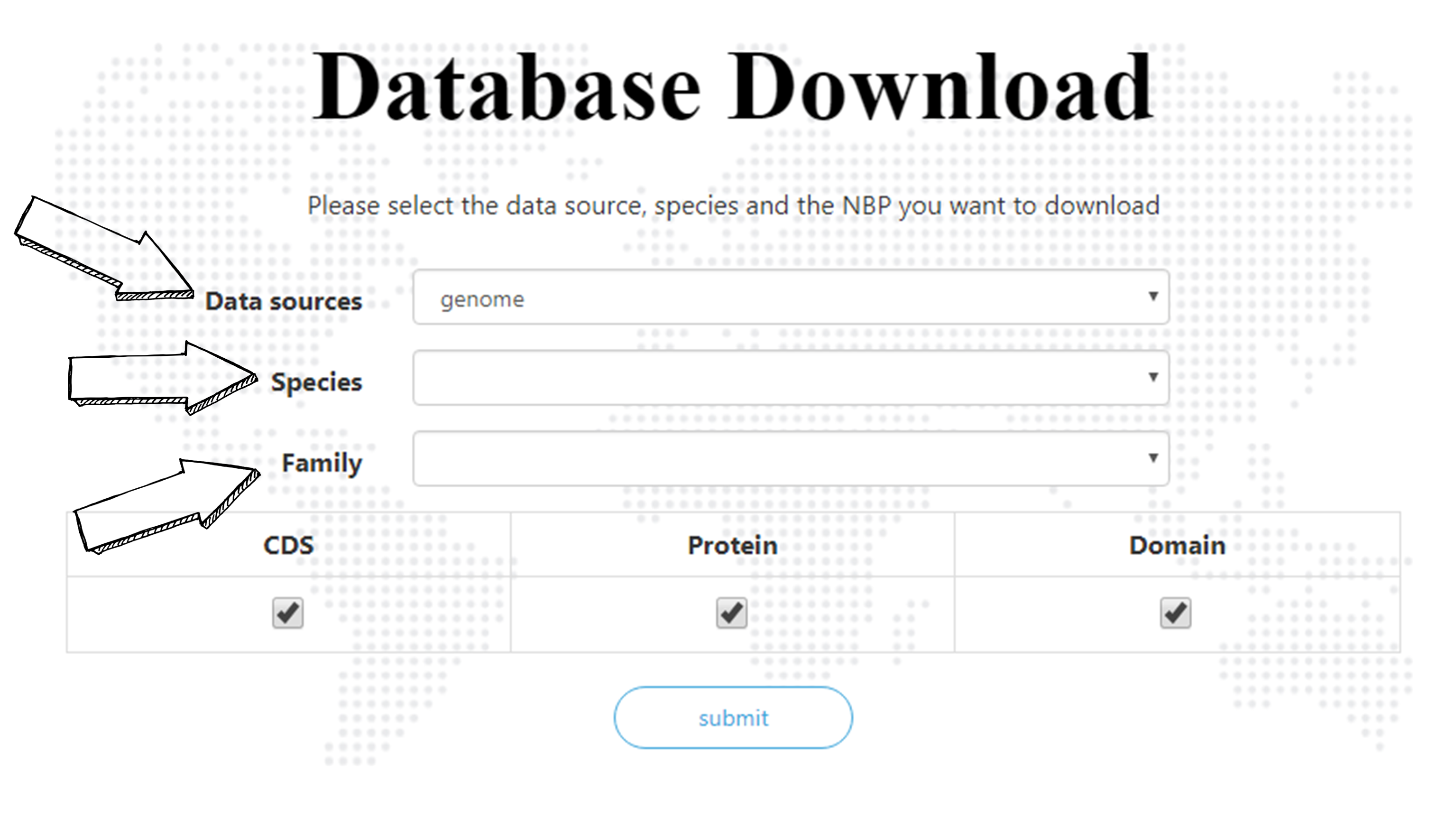
Download Database
ENPD allows users to download the sequences of NBPs. First please select the source of data. Users can select the species and NBP family they want and download CDS, protein, or/and domain sequences. Users can also download sequences of a certain NBP family in all species by leaving the “Species” blank and selecting a family, download sequences of all NBP families in a certain species by leaving the “Family” blank and selecting a species, or download all sequences by leaving both blank.
Species browser
ENPD allows users to browse through the NBP statistics and taxonomical information through the species browser function.
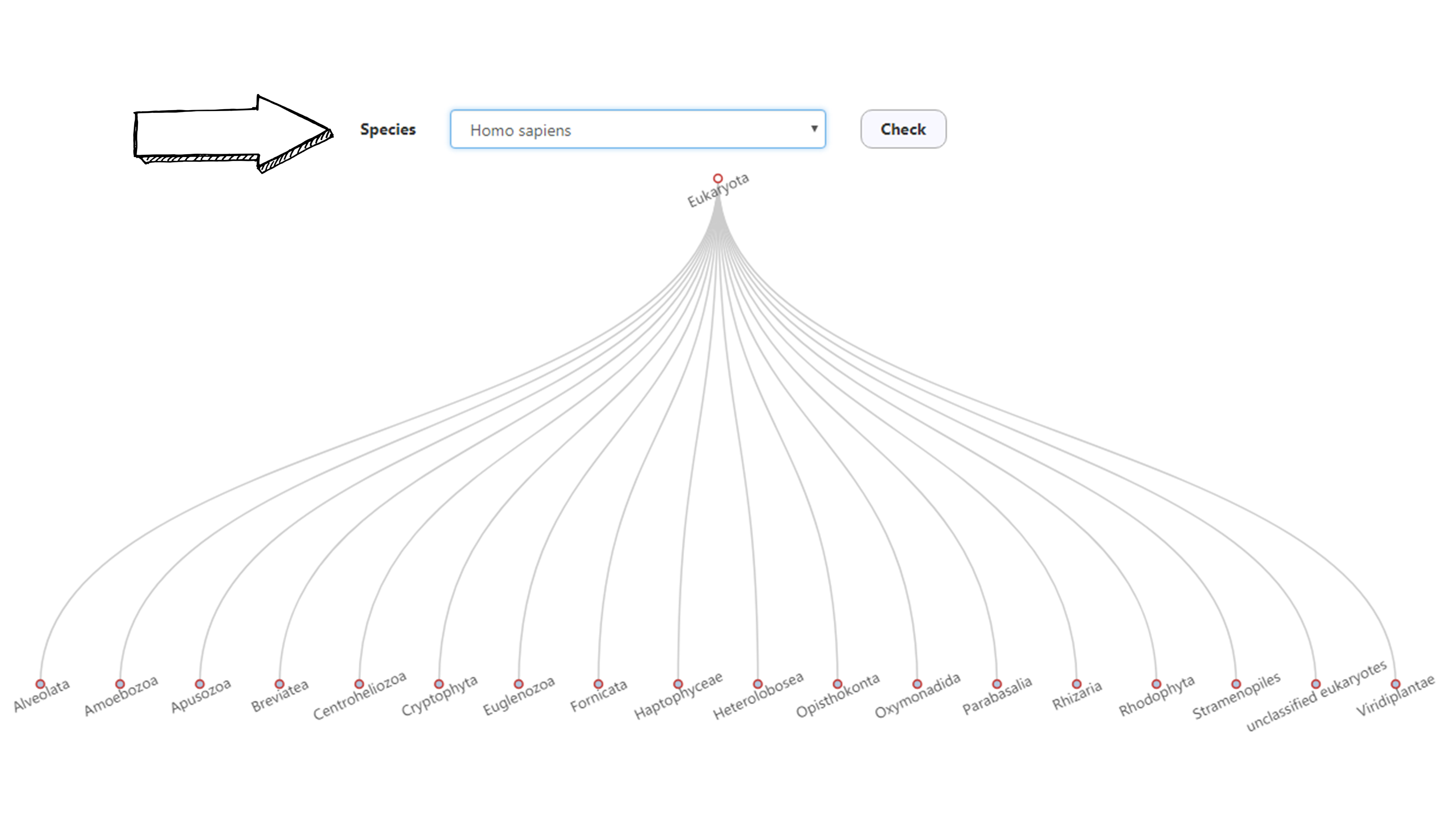
Choose the specie of your interest, click check. It shall return with a NBP statistic table and the full taxonomic information of the species you selected.
