
Eukaryotic Nucleic acid binding Protein Database (ENPD) is an online library of nucleic acid binding proteins (NBPs) and their functional information. Nucleic acid binding proteins (NBPs), i.e., DNA binding proteins (DBPs), RNA-binding proteins (RBPs) as well as DNA and RNA binding proteins (DRBPs) are capable of regulating genes by interacting with DNA, RNA or both respectively. They are highly cooperated with each other and perform vital functions in every stage of gene regulation (1-3). To enhance and facilitate our understanding towards gene regulation mediated by nucleic acid binding proteins, the Nucleic acid binding proteins (NBPs) repertoires of various eukaryotic organisms- Eukaryotic Nucleic acid binding Protein Database have been constructed based on manual curation and a new developed pipeline by analyzing sequenced transcriptomes and genomes. From transcriptomes and genomes available for around 1770 and 790 eukaryotic species respectively, 662 NBP families with more than 2866174 of NBPs were predicted with inferred binding motifs for a total number of 2423 species, constituting the largest NBP database that includes DBPs, RBPs, and DRBPs.
Nucleic Binding Protein statistic summary from 6 model organisms
| Species | DRBP | RBP | TF | NBP number |
|---|---|---|---|---|
| Homo sapiens | 2736 | 383 | 1635 | 4754 |
| Mus musculus | 3727 | 927 | 3135 | 7789 |
| Drosophila melanogaster | 1024 | 311 | 1002 | 2337 |
| Caenorhabditis elegans | 390 | 139 | 807 | 1336 |
| Gallus gallus | 2025 | 728 | 1981 | 4734 |
| Danio retio | 4917 | 901 | 3251 | 9069 |
 Download Picture
Download Picture
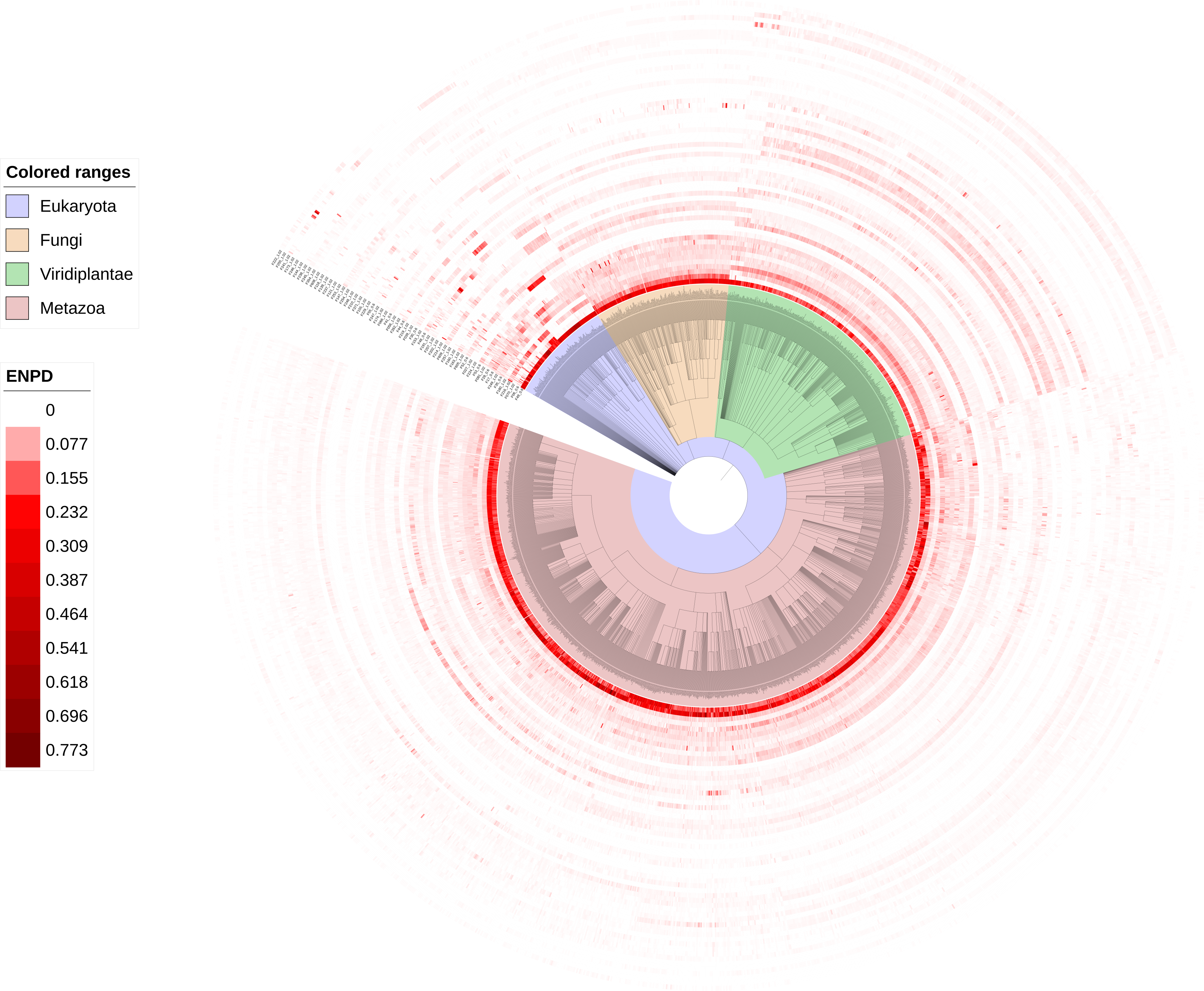
NBP families compared within eukaryotic lineages. Colors in the figure show the percentage of NBPs in each NBP family over all predicted NBPs in a species. NBP families were found to be expanded or absent in different lineages. For example, family F49- RRM was found to be prevalence in all eukaryotic lineages while F06- C2H2 ZF was found expanded in animals and fungi but not in plants. This database covers evolutionarily important lineages such as Alismatales (basal monocots), early-diverging eudicots (primitive dicots), Palaeoptera (an ancestral group of winged insects), Bivalvia (mollusks with shells divided into two halves) and Cephalopoda (the lineage containing squid and octopus), none of which are recorded in the previously NBP databases.
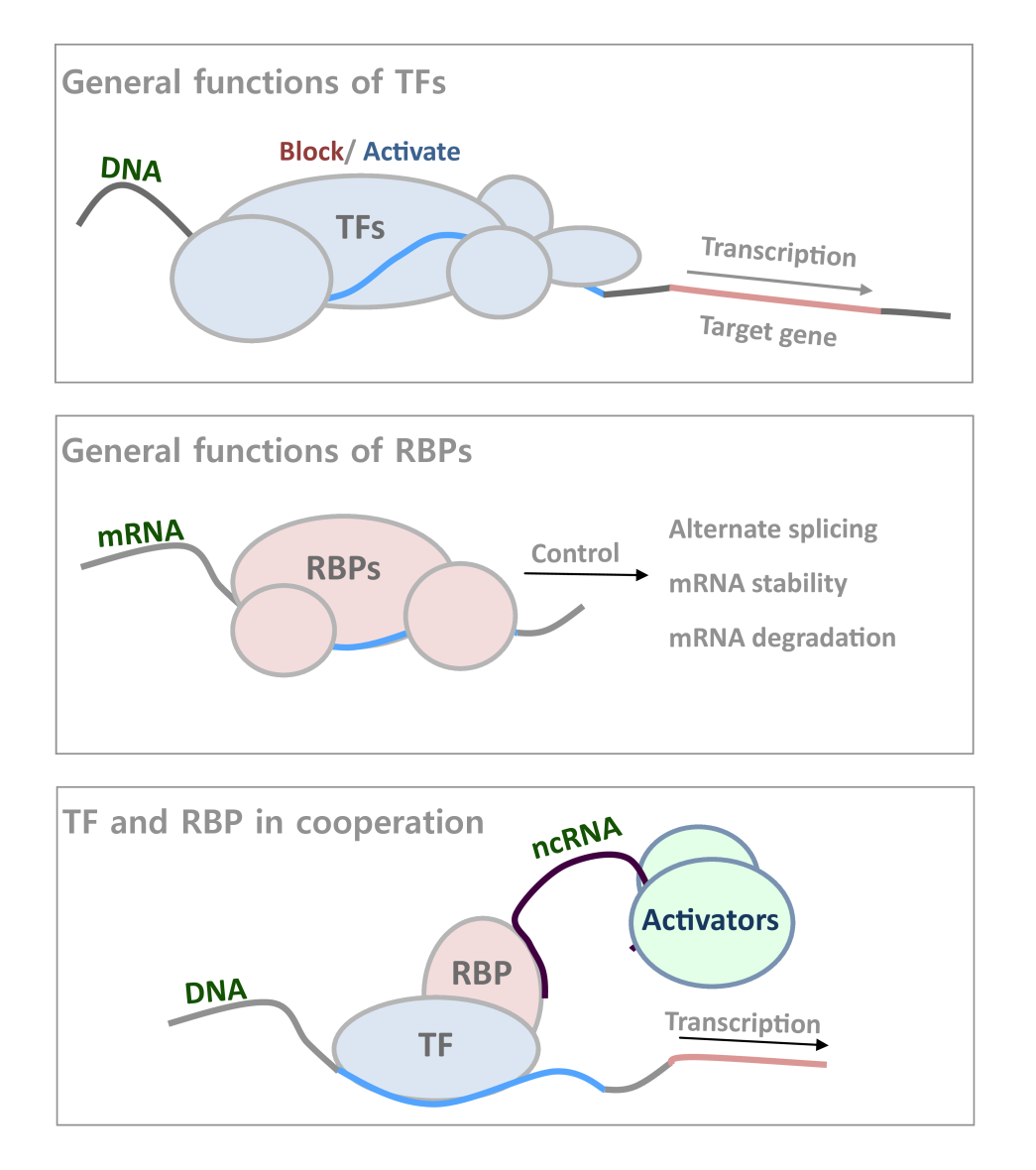
General Functions of TFs and RBPs. Transcription factors (TFs) and RNA-binding proteins (RBPs) are involved in every stage of gene regulation and are important for most if not all biological processes through interactions with DNAs and RNAs, controlling cell identity and pathological status. TFs usually bind to DNA regulatory elements of a gene, and regulate its transcription activity; while RBPs have diverse functions related to ncRNA mediated gene regulation as well as mRNA regulation, including alternate splicing, mRNA stability control, and degradation (1,2). The two types of protein can also cooperate together recruiting lncRNA for further activators recruitment(4).
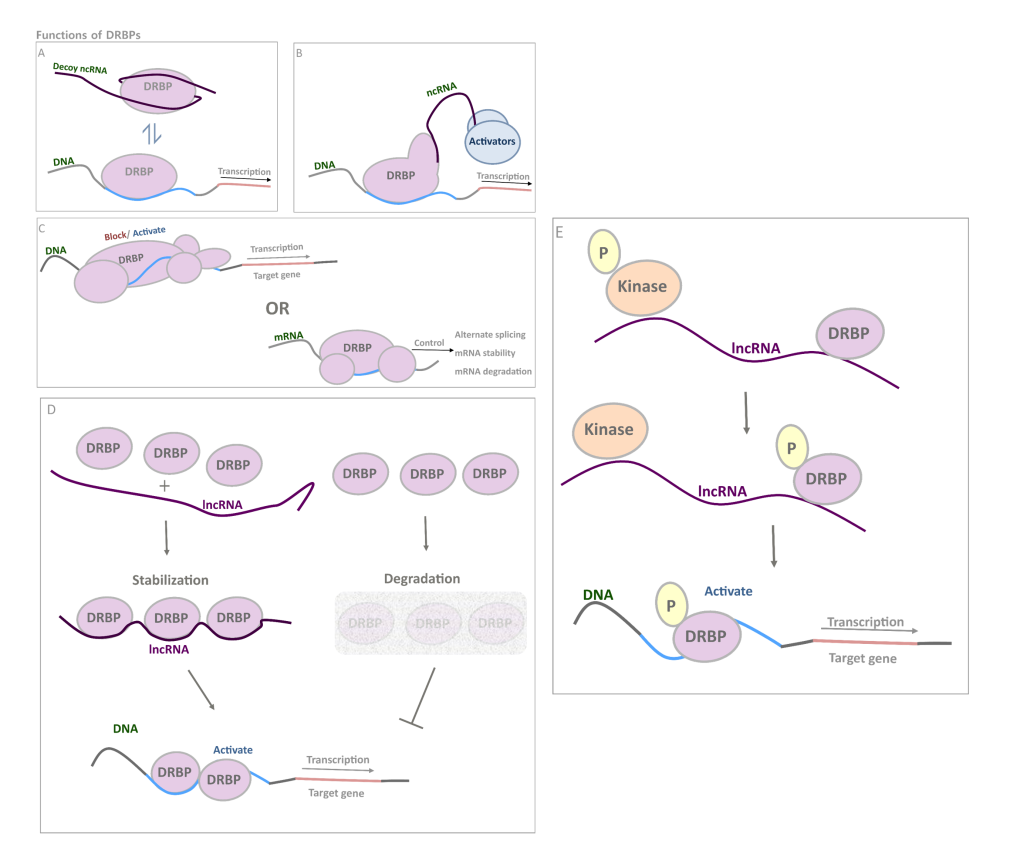
Functions of DRBPs. DRBPs have a wide range of distinct functions; (A) they could function as a typical TF with add-on abilities such as interacting with ncRNA decoys(5); (B) recruitment of ncRNA with co-activator function(6-10); (C) regulating mRNAs and DNA regulatory elements just as typical RBPs and TFs(11-14); (D) interact with lncRNA for stability enhancement, thus increase its own abundance and further activate downstream genes(15); (E) alter its phosphorylation status by the mediation of lncRNA, enhancing its gene activating activity(16).
Learn More Start Search Contact Us Related Links
REFERENCE
1. Glisovic, T., Bachorik, J.L., Yong, J. and Dreyfuss, G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett, 582, 1977-1986.
2. Latchman, D.S. (1997) Transcription factors: an overview. Int J Biochem Cell Biol, 29, 1305-1312.
3. Hudson, W.H. and Ortlund, E.A. (2014) The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol, 15, 749-760.
4. Marchese, F.P., Raimondi, I. and Huarte, M. (2017) The multidimensional mechanisms of long noncoding RNA function. Genome Biol, 18, 206.
5. Kino, T., Hurt, D.E., Ichijo, T., Nader, N. and Chrousos, G.P. (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal, 3, ra8.
6. Xu, B., Yang, W.H., Gerin, I., Hu, C.D., Hammer, G.D. and Koenig, R.J. (2009) Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol, 29, 1719-1734.
7. Deblois, G. and Giguere, V. (2003) Ligand-independent coactivation of ERalpha AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J Steroid Biochem Mol Biol, 85, 123-131.
8. Xu, B. and Koenig, R.J. (2004) An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J Biol Chem, 279, 33051-33056.
9. Poon, M.M. and Chen, L. (2008) Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A, 105, 20303-20308.
10. Zhao, X., Patton, J.R., Davis, S.L., Florence, B., Ames, S.J. and Spanjaard, R.A. (2004) Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell, 15, 549-558.
11. Auphan, N., DiDonato, J.A., Rosette, C., Helmberg, A. and Karin, M. (1995) Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science, 270, 286-290.
12. Surjit, M., Ganti, K.P., Mukherji, A., Ye, T., Hua, G., Metzger, D., Li, M. and Chambon, P. (2011) Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell, 145, 224-241.
13. Jonat, C., Rahmsdorf, H.J., Park, K.K., Cato, A.C., Gebel, S., Ponta, H. and Herrlich, P. (1990) Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell, 62, 1189-1204.
14. Dhawan, L., Liu, B., Blaxall, B.C. and Taubman, M.B. (2007) A novel role for the glucocorticoid receptor in the regulation of monocyte chemoattractant protein-1 mRNA stability. J Biol Chem, 282, 10146-10152.
15. Wei, M.M., Zhou, Y.C., Wen, Z.S., Zhou, B., Huang, Y.C., Wang, G.Z., Zhao, X.C., Pan, H.L., Qu, L.W., Zhang, J. et al. (2016) Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget, 7, 59556-59571.
16. Li, D., Liu, X., Zhou, J., Hu, J., Zhang, D., Liu, J., Qiao, Y. and Zhan, Q. (2017) Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology, 65, 1612-1627.